Following transport to the cell surface, secreed proteins involved in such critical cell processes as surface adhesion and mating are anchored to the cell surface. A better understanding of the molecular machineries involved in protein anchoring will facilitate the development of effective approaches for interfering with surface anchoring of such specific targets as virulence factors.
We have discovered that the N-terminal region of H. volcanii Tat substrates often contain a lipobox, a motif that is modified by the covalent attachment of a lipid to an invariant cysteine. Although archaea do not have homologues of the bacterial enzymes involved in lipid modification of lipobox-containing proteins, we have identified two candidates that might be involved in this process, as they are found in all archaea that express proteins containing a lipobox but not found in other archaea.
Recently, we confirmed that some archaea, including H. volcanii, also employ another type of protein anchoring mechanism that is mediated through a lipid-modified amino acid residue. In this case, an enzyme known as the archaeosortase processes, and attaches a lipid to, the C-terminus of substrates containing a PGF motif. One such substrate, the S-Layer Glycoprotein (SLG), perhaps the best studied of all archaeal proteins, was long believed to be anchored to the cell membrane by intercalation of a protein segment.
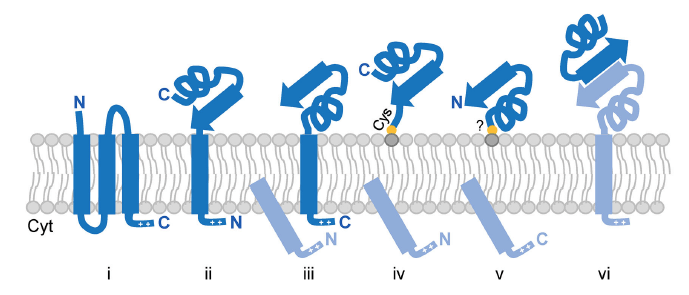
Fig. 1. Anchoring strategies of archaeal surface proteins. Proteins can be anchored via multiple TM domains (i), single N- or C-terminal TM domains (ii and iii, respectively); N- or C-terminal covalent lipid interactions (iv and v, respectively); or interactions with other surface-anchored proteins (vi). Cleaved signal peptides and interacting surface proteins are colored in light blue.
We are now characterizing additional archaeosortase substrates, including several proteins that contain a low-complexity domain, a type of domain often found in proteins involved in mediating surface adhesion or cell-cell interactions.
Selected publications
- F. Abdul-Halim, S. Schulze, A. DiLucido, F. Pfeiffer, A. W. Bisson Filho, M. Pohlschroder 2020. Lipid Anchoring of Archaeosortase Substrates and Midcell Growth in Haloarchaea. mBio 11(2).
- Pohlschroder M. and S. Schulze. 2018. Haloferax volcanii. Microbe of the Month. Trends in Microbiology. Trends in Microbiol. 27:86-87.
- Pohlschroder, M. and S. Albers. 2018. Editorial: Editorial for thematic issue on Archaea. FEMS Microbiol. Rev. 42: 719-720.
- Pohlschroder M., S. Schulze, F. Pfeiffer and MF Abdul Halim. 2018. Archaeal Cell Surface Biogenesis. 2018. FEMS Microbiology Reviews. 42: 694-717.
- Abdul Halim MF, R, Rodriguez, JD Stoltzfus, IG Duggin, Pohlschroder. 2018. Conserved residues are critical for Haloferax volcanii archaeosortase catalytic activity: Implications for convergent evolution of the catalytic mechanisms of non-homologous sortases from archaea and bacteria. Mol. Micro. 108:276-287.
- Abdul Halim, MF, J. Stolzfus, S. Schulze, M. Hippler and Pohlschroder. 2017. ArtA-dependent processing of a Tat substrate containing a conserved tripartite structure that is not localized at the C-terminus. J. Bacteriol. 199 e00802-16
- Halim, MFA, J. Stolzfus, S. Schulze, M. Hippler and M. Pohlschroder. 2016. ArtA-dependent processing of a Tat substrate containing a conserved tripartite structure that is not localized at the C-terminus. J. Bacteriol. In press.
- Abdul Halim, M. F, F. Pfeiffer, J. Zou, A. Frisch, D. Haft, S. Wu, N. Tolic, H. Brewer, S. H. Payne, L. Paša-Tolic, and M. Pohlschroder. 2013. Haloferax volcanii archaeosortase is required for motility, mating and C-terminal processing of the S-layer glycoprotein. Mol. Microbiol. 85:1164-1175.
- Halim, F., K. Karch, Y. Zhou, B. Garcia and M. Pohlschroder. 2016. Permuting the PGF-CTERM signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 198:808-815.
- Storf S., F. Pfeiffer, K. Dilks, Z. Chen, Imam, and M. Pohlschroder. 2010. Mutational and bioinformatic analysis of haloarchaeal lipoproteins. Archaea. 2010 Article ID 75410.
- Gimenez M. I., K. Dilks, and M. Pohlschroder. 2007. Haloferax volcanii twin arginine translocation substrates include soluble secreted, C-terminally anchored and lipoproteins. Mol. Microbiol. 66:1597-1602.
- Storf S., F. Pfeiffer, K. Dilks, Z. Chen, Imam, and M. Pohlschroder. 2010. Mutational and bioinformatic analysis of haloarchaeal lipoproteins. Archaea. 2010 Article ID 75410.
