2025
Lin SY, Futeran HR, Cruga BN, Santiago-Frangos A, Levine MT*.: Rapid compensatory evolution within a multiprotein complex preserves telomere integrity. Science.

Will’s paper (Henriques et al., 2025) below was featured in this wonderful perspective piece “Capture first, then deliver!” by Iktae Kim and Jeong-Yong Suh!

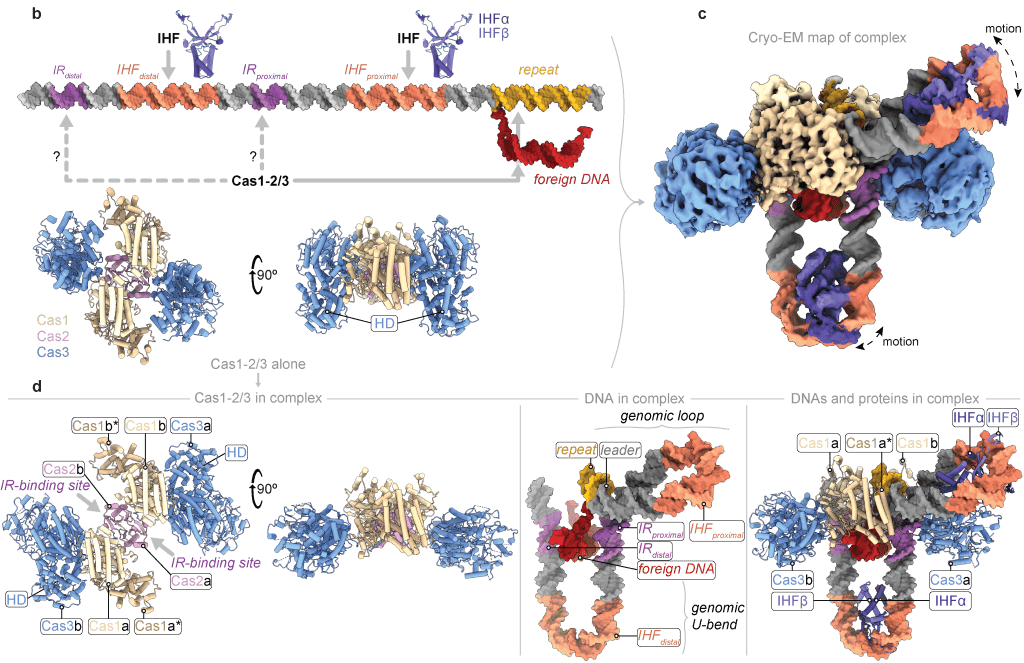
Henriques WS, Bowman J, Hall LN, Gauvin CC, Wei H, Kuang H, Zimanyi CM, Eng ET, Santiago-Frangos A, Wiedenheft B*.: Structures reveal how the Cas1-2/3 integrase captures, delivers, and integrates foreign DNA into CRISPR loci.
Structure.

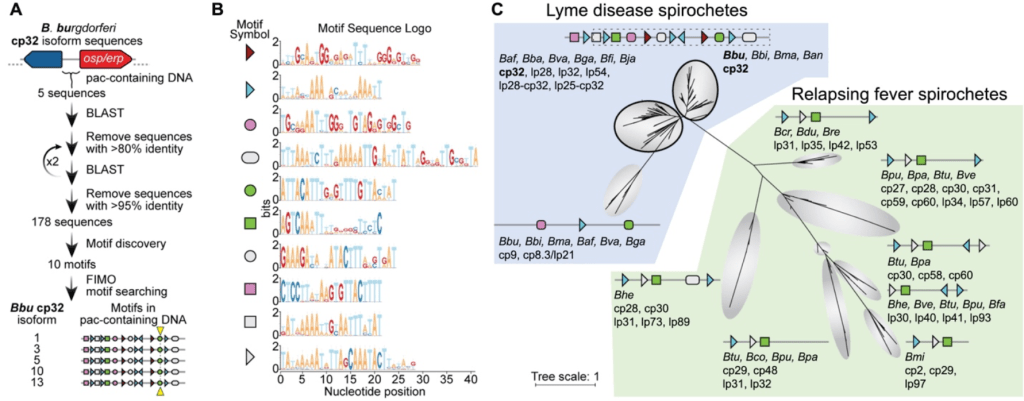
Buyukyoruk M, Krishna P, Santiago-Frangos A*, Wiedenheft B*.: Discovery of Diverse CRISPR Leader Motifs, Putative Functions, and Applications for Enhanced CRISPR Detection and Subtype Annotation. The CRISPR Journal.

2024
Faith DR, Kinnersley M, Brooks DM, Drecktrah D, Hall LS, Luo E, Santiago-Frangos A, Wachter J, Samuels DS, Secor PR.: Characterization and genomic analysis of the Lyme disease spirochete bacteriophage phiBB-1. PLOS Pathogens.
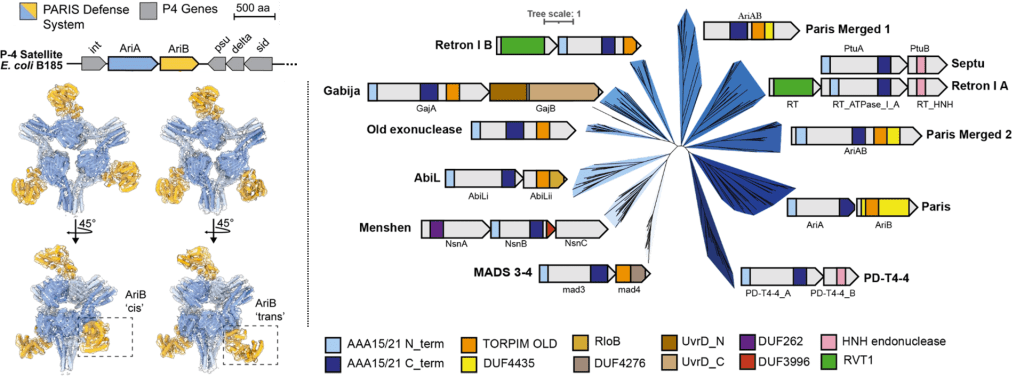
Burman N, Belukhina S, Depardieu F, Wilkinson RA, Skutel M, Santiago-Frangos A, Graham AB, Livenskyi A, Chechenina A, Morozova N, Zahl T, Henriques WS, Buyukyoruk M, Rouillon C, Shyrokova L, Kurata T, Hauryliuk V, Severinov K, Groseille J, Thierry A, Koszul R, Tesson F, Bernheim A, Bikard A*, Wiedenheft B*, Isaev A*.: Viral proteins activate PARIS-mediated tRNA degradation and viral tRNAs rescue infection. bioRxiv.
2023 – Prior to the University of Pennsylvania –
Santiago-Frangos A, Henriques WS, Wiegand T, Gauvin CC, Buyukyoruk M, Graham AB, Wilkinson RA, Triem L, Neselu K, Eng E, Lander GC, Wiedenheft B*.: Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays. Nature Structural & Molecular Biology.
Wiegand T, Wilkinson R, Santiago-Frangos A, Lynes M, Hatzenpichler R, Wiedenheft B*.: Functional and Phylogenetic Diversity of Cas10 Proteins. The CRISPR Journal.
2022
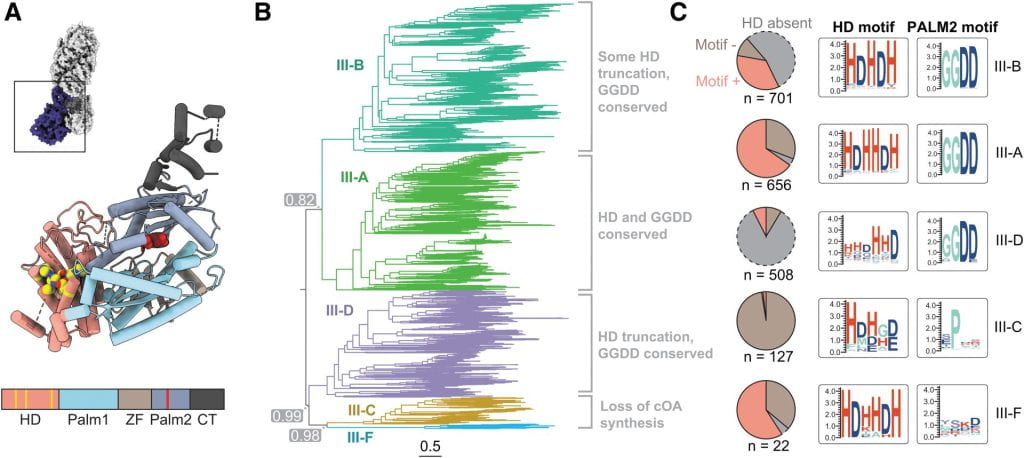
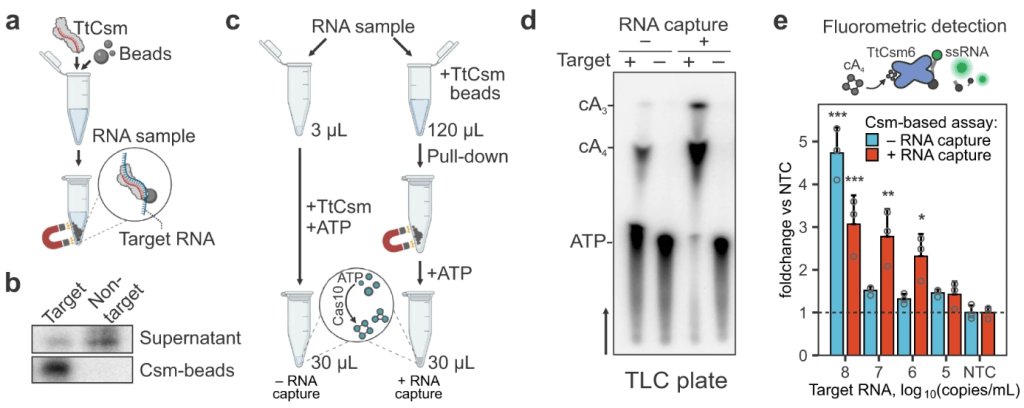
Nemudraia A, Nemudryi A, Buyukyoruk M, Scherffius AM, Zahl TR, Wiegand T, Pandey S, Nichols JE, Hall L, McVey A, Lee HH, Wilkinson RA, Snyder LR, Jones JD, Koutmou KS, Santiago‐Frangos A*, Wiedenheft B*.: Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic. Nature Communications
Patent
Nucleic acid detection using type iii crispr complex.
Blake A. Wiedenheft, Andrew Santiago-Frangos, Anna A. Nemudraia, Artem A. Nemudryi.
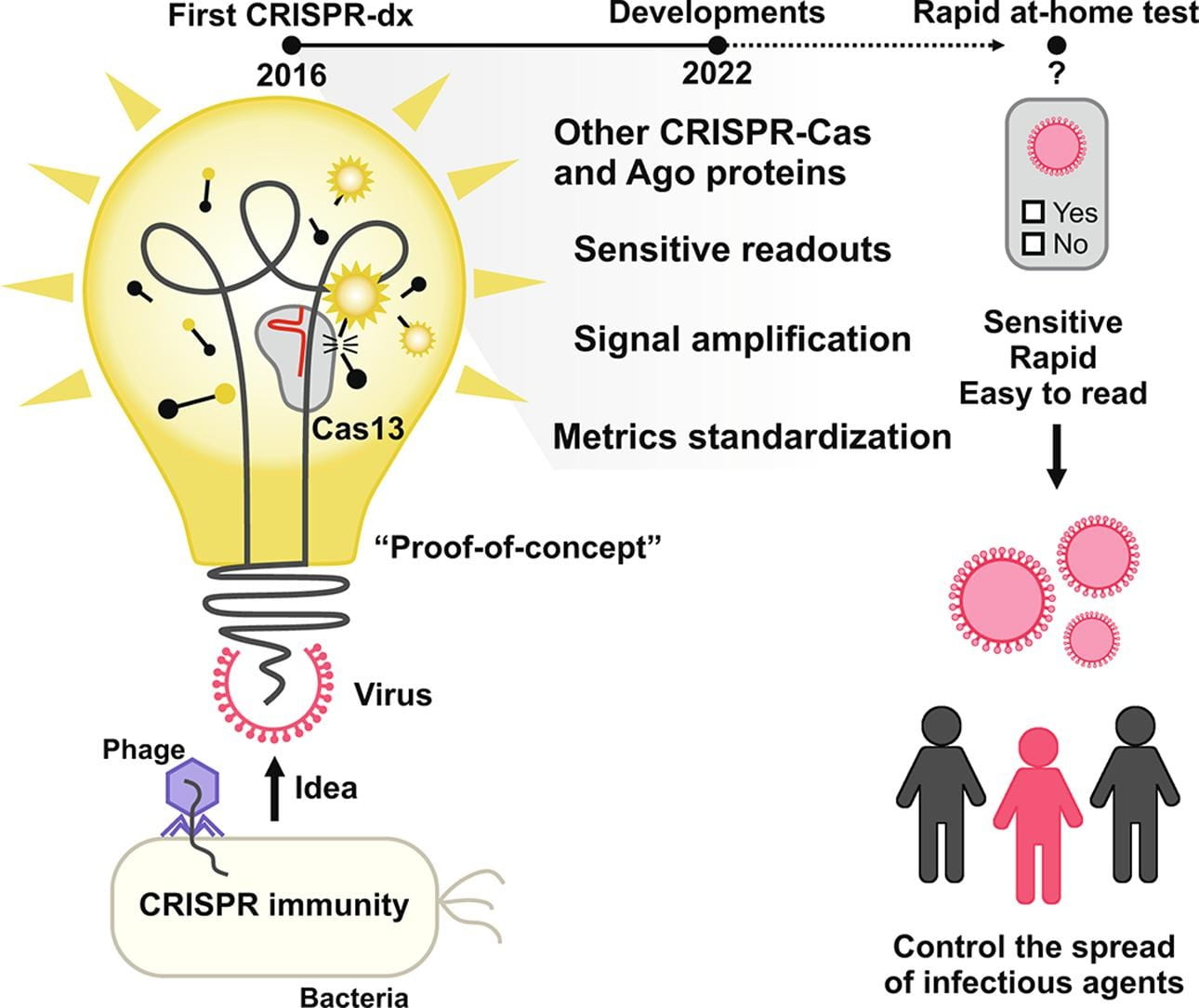
Santiago‐Frangos A, Nemudryi A, Nemudraia A, Wiegand T, Nichols JE, Krishna P, Scherffius AM, Zahl TR, Wilkinson RA, Wiedenheft B*.: CRISPR-Cas, Argonaute proteins and the emerging landscape of amplification-free diagnostics. Methods.
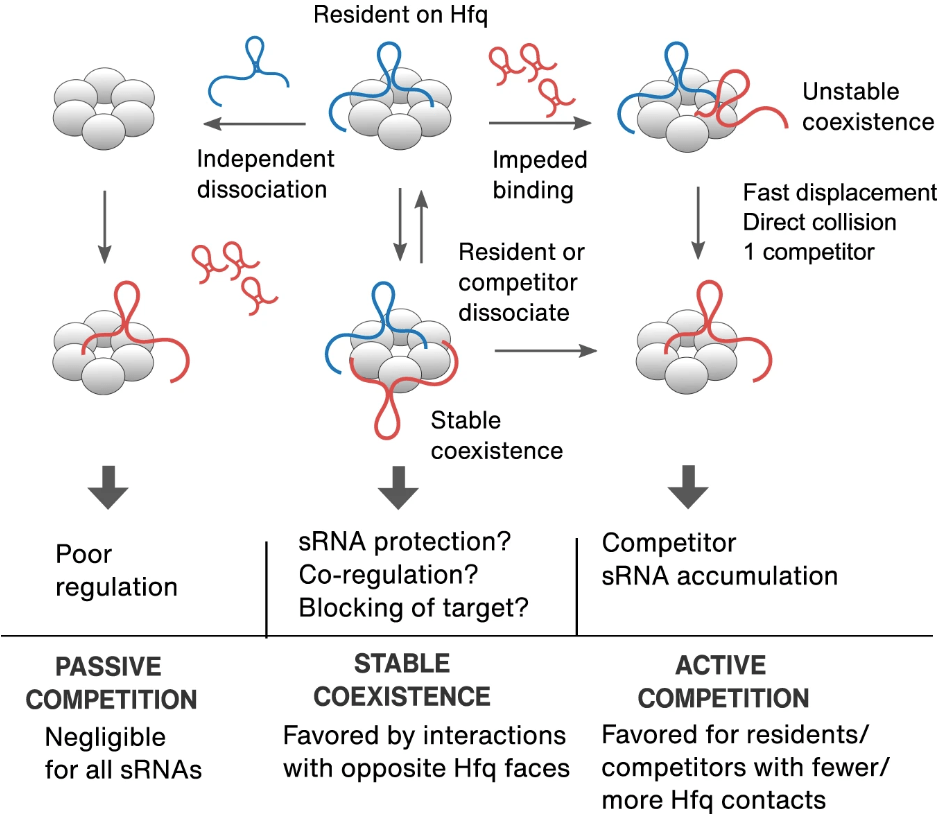
Roca J, Santiago‐Frangos A, Woodson SA*.: Diversity of bacterial small RNAs drives competitive strategies for a mutual chaperone. Nature Communications.
2021
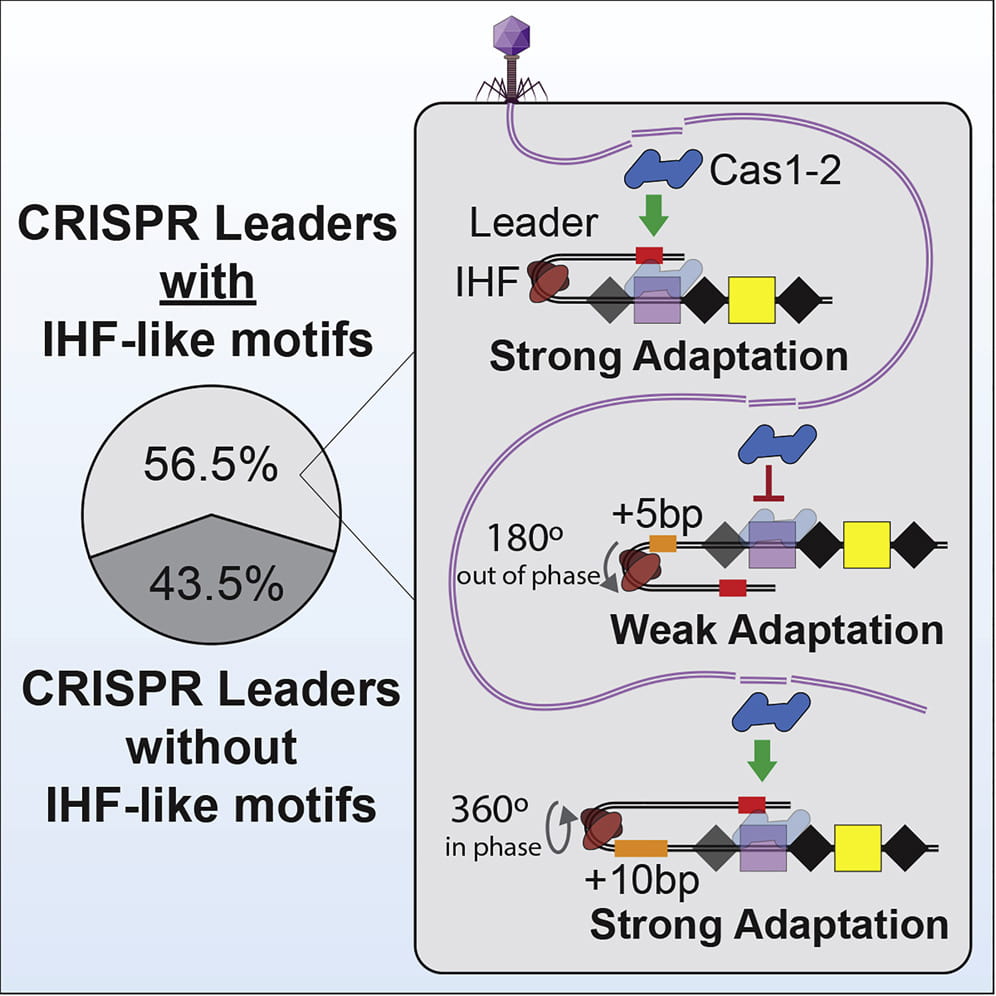
Santiago‐Frangos A, Buyukyoruk M, Wiegand T, Krishna P, Wiedenheft B*.: Distribution and phasing of sequence motifs that facilitate CRISPR adaptation. Current Biology.
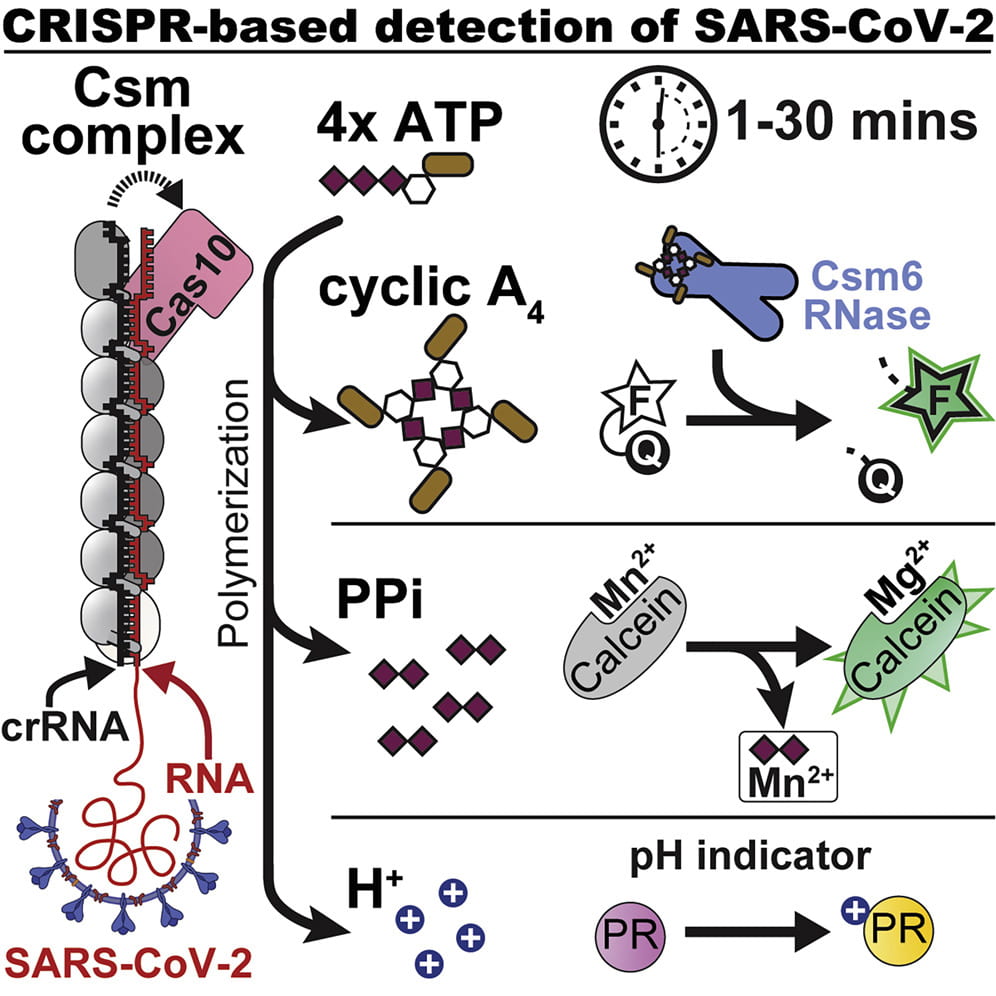
Santiago-Frangos A, Hall LN, Nemudraia A, Nemudryi A, Krishna P, Wiegand T, Wilkinson RA, Snyder DT, Hedges JF, Cicha C, Lee HH, Graham A, Jutila MA, Taylor MP, Wiedenheft B*.: Intrinsic signal amplification by type III CRISPR-Cas systems provides a sequence-specific SARS-CoV-2 diagnostic. Cell Reports Medicine.
Patent
Engineered crispr-cas systems and methods for sensitive and specific diagnostics.
Blake A. Wiedenheft, Andrew Santiago-Frangos, Anna A. Nemudraia, Artem A. Nemudryi.
2020
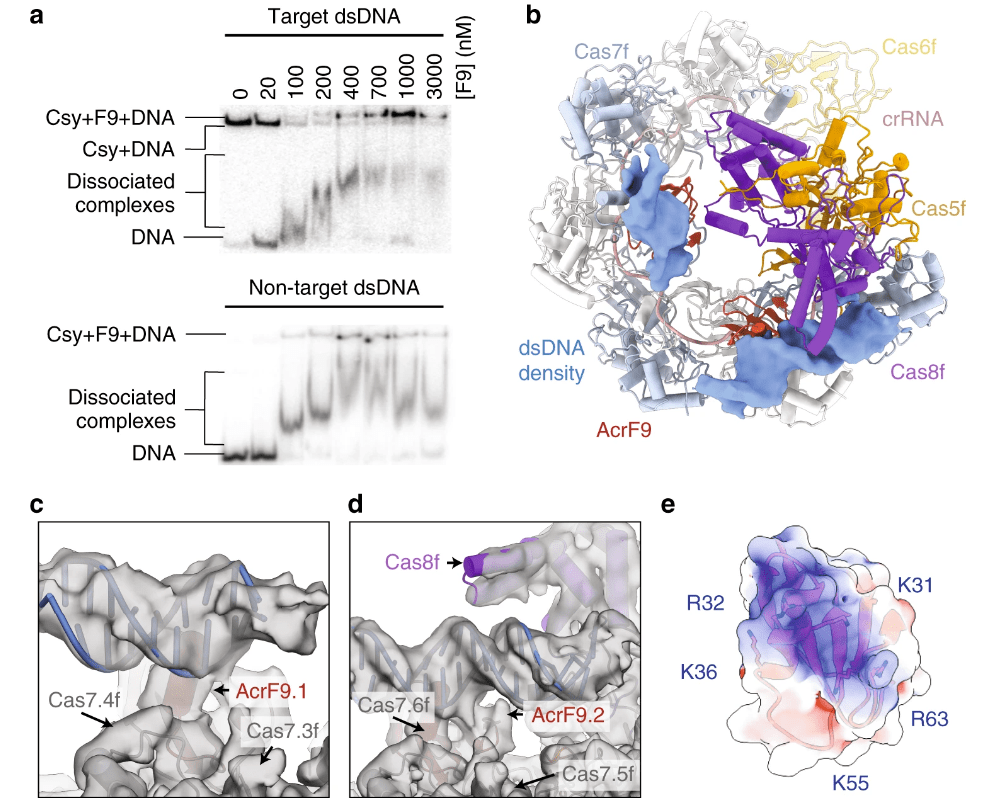
Hirschi M †, Lu WT †, Santiago-Frangos A, Wilkinson, R, Golden SM, Davidson AR, Lander GC, Wiedenheft B*.: AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex. Nature Communications.
Panja S, Małecka EM, Santiago-Frangos A, Woodson SA*.: Quantitative analysis of RNA chaperone activity by native gel electrophoresis and fluorescence spectroscopy. RNA chaperones: Methods and Protocols.
2019
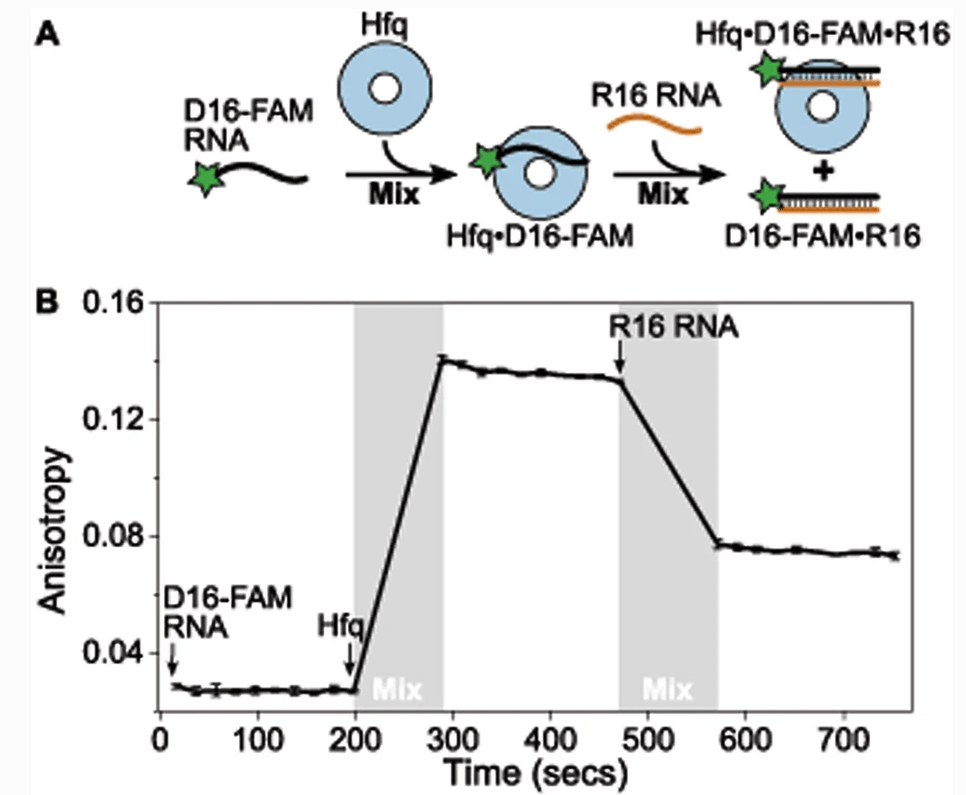
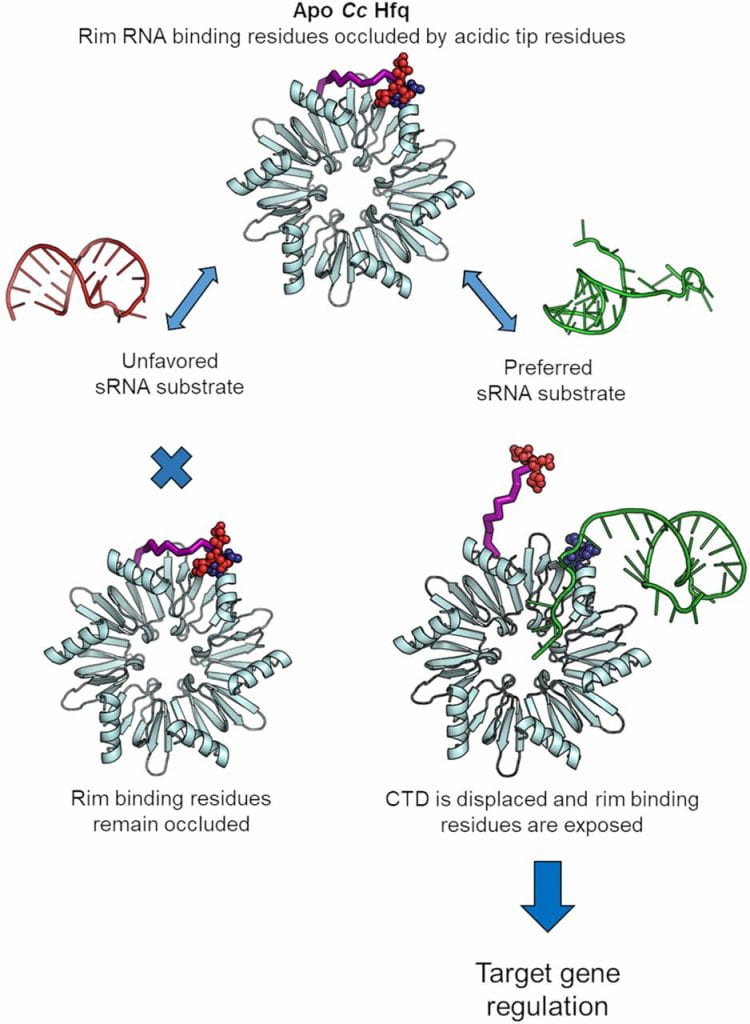
Santiago-Frangos A †, Fröhlich KS †, Jeliazkov JR, Małecka EM, Marino G, Gray JJ, Luisi BF*, Woodson SA*, Hardwick SW*.: Caulobacter Crescentus Hfq Structure Reveals a Conserved Mechanism of RNA Annealing Regulation. Proceedings of the National Academy of Sciences.
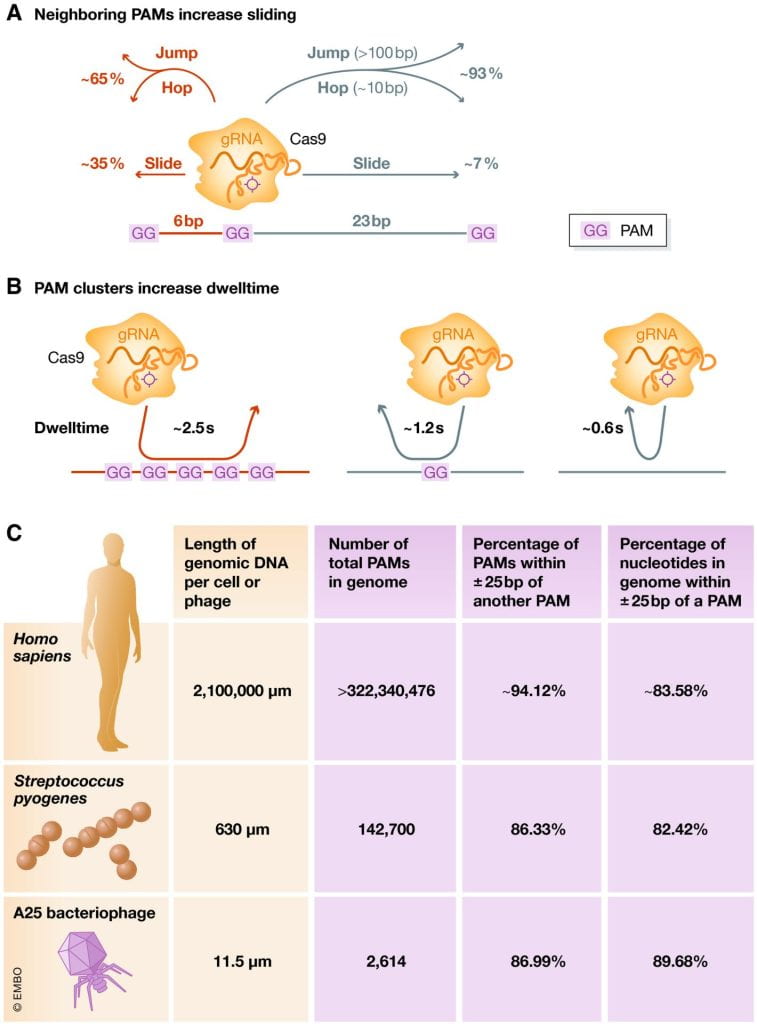
Santiago‐Frangos A, Wiegand T, Wiedenheft B.: Cas9 slide‐and‐seek for phage defense and genome engineering. EMBO Journal.
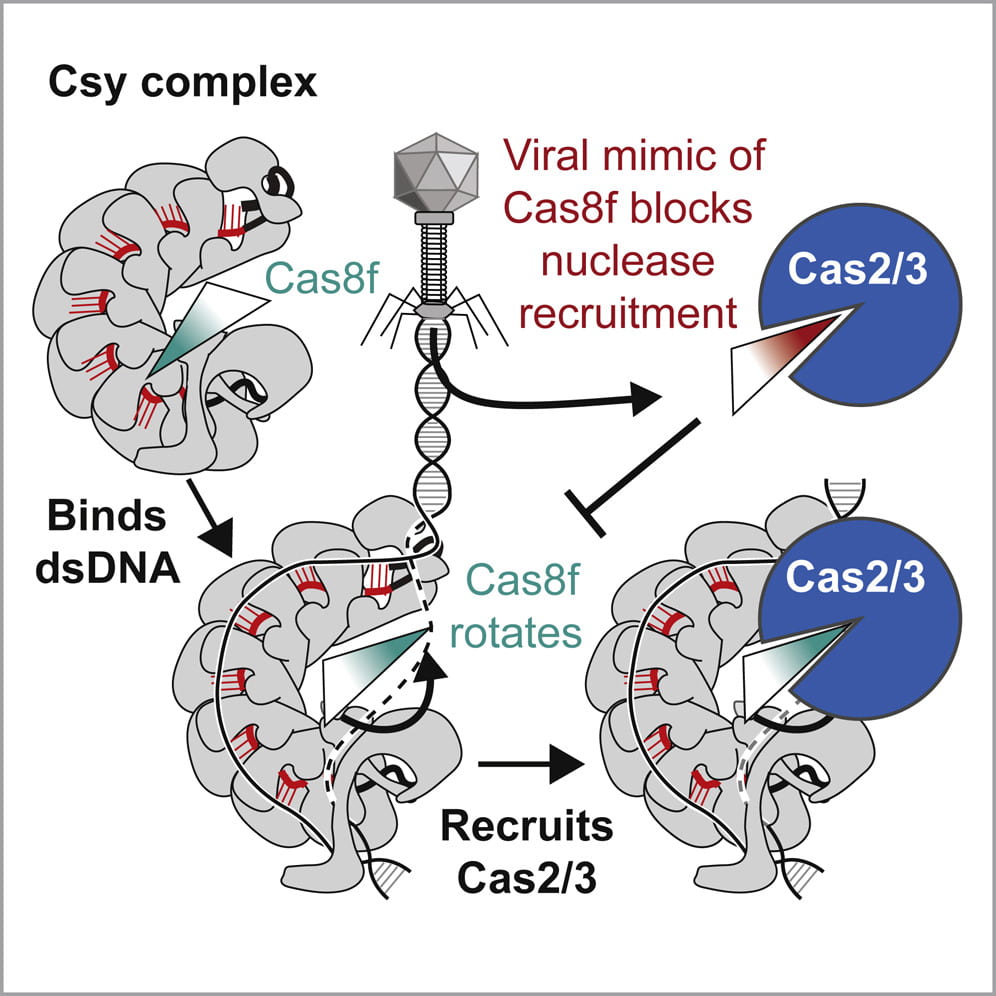
Rollins M.F. †, Chowdhury S. †, Carter J., Golden S.M., Miettinen H.M., Santiago-Frangos A. Faith D, Lawrence CM, Lander GC*, Wiedenheft B*.: Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry. Molecular Cell.
2018
Woodson SA*, Panja S, Santiago-Frangos A. Proteins That Chaperone RNA Regulation. Microbiology Spectrum.
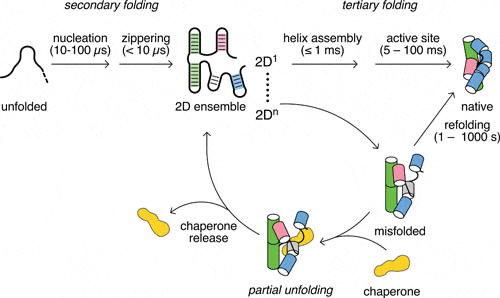
Santiago-Frangos A, & Woodson, SA*. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdisciplinary Reviews: RNA.
2017
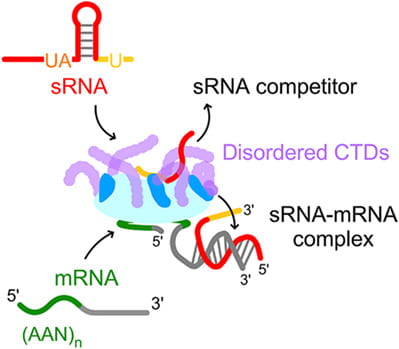
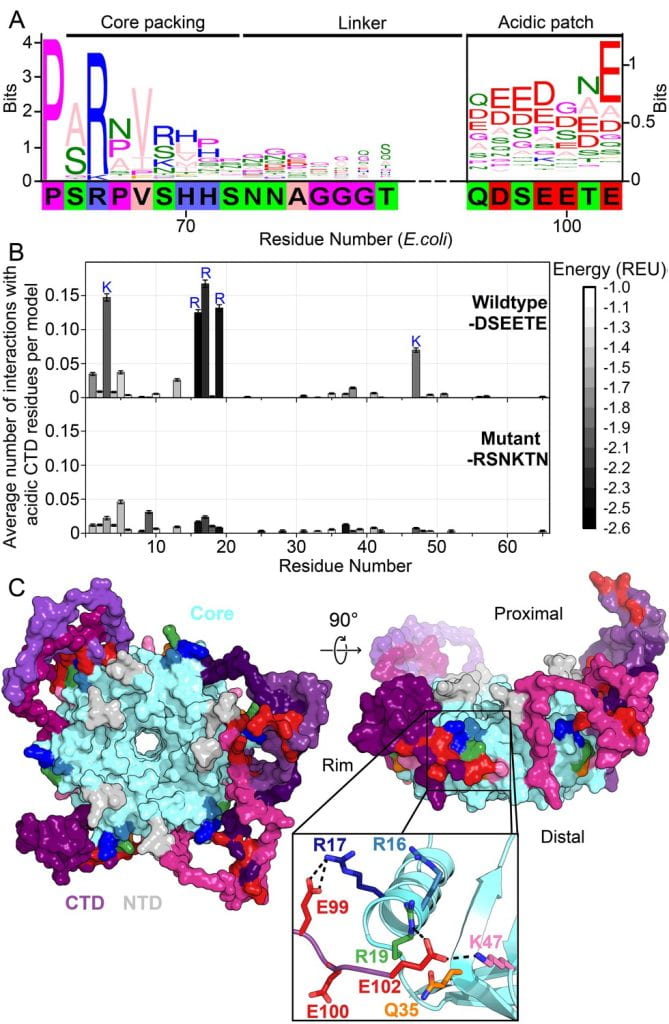
Santiago-Frangos A, Jeliazko JR, Gray JJ, Woodson SA*. Acidic C-terminal domains autoregulate the RNA chaperone Hfq. Elife.
2016
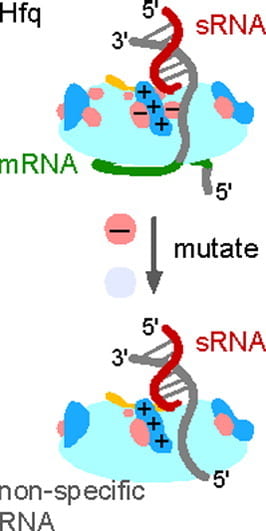
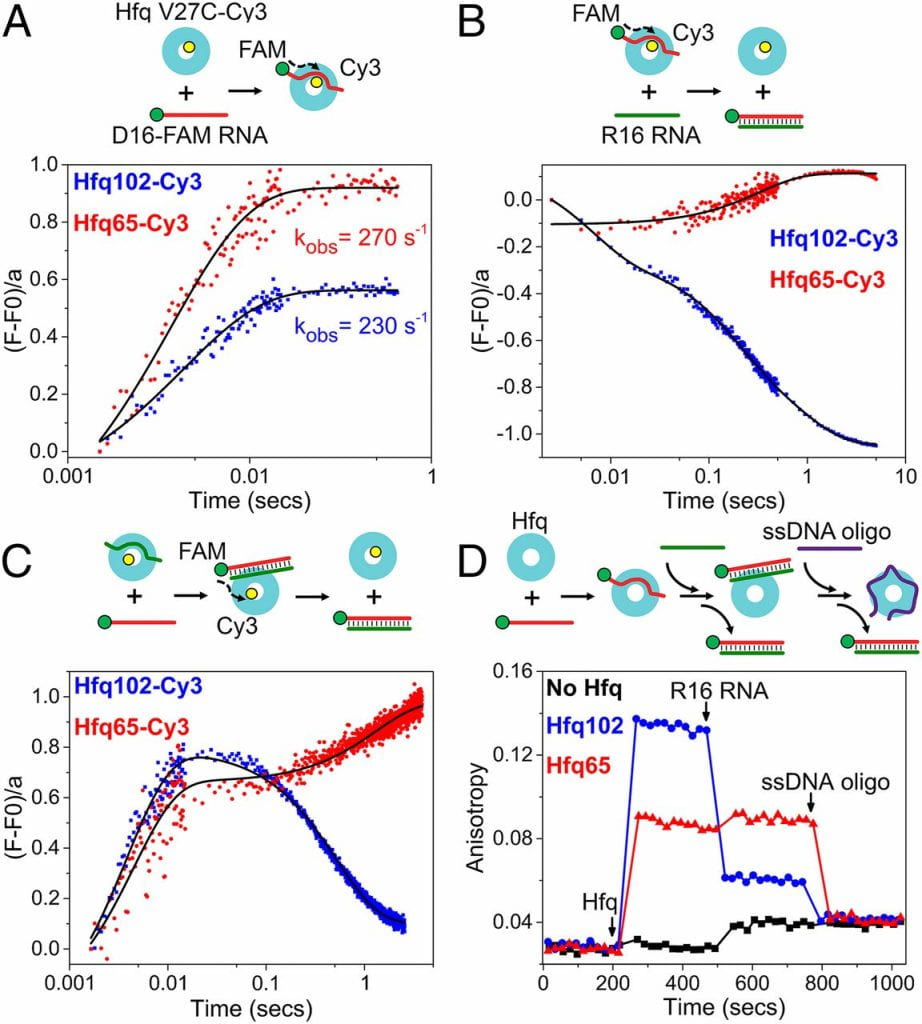
Santiago-Frangos A, Kavita K, Schu DJ, Gottesman S, Woodson SA. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proceedings of the National Academy of Sciences.
2015
Panja S †, Santiago-Frangos A †, Schu DJ, Gottesman S, Woodson SA. Acidic residues in the Hfq chaperone increase the selectivity of sRNA binding and annealing. Journal of Molecular Biology.