MISSION
Combine computational biology, biochemistry, microbiology, and structural biology to determine how bacteria and archaea sense and respond to phage infections. In addition to satisfying an innate curiosity for how the world around us functions, we’ll repurpose these mechanistic insights for medical or biotech applications.
Mechanisms and applications of diverse CRISPR integration complexes
Vertebrates, bacteria, and archaea have domesticated transposases (e.g., RAG1 and Cas1) for adaptive immunity. Transposases, integrases and recombinases often co-opt additional DNA-bending proteins (e.g., IHF, HU, H-NS, or HMGB1) that facilitate DNA integration and excision. However, the structural role of DNA folding during this mobilization of DNA remains largely enigmatic.

Bacteria and archaea acquire resistance to viruses and plasmids by integrating fragments of foreign DNA into the first repeat of a CRISPR array. Preferential integration at the first CRISPR repeat ensures efficient transcription and processing of CRISPR RNAs that target the most recently encountered genetic parasites.
We recently determined a 560 kDa integration complex structure that explains how Pseudomonas aeruginosa Cas (Cas1-2/3) and non-Cas proteins (IHF) bind conserved DNA sequence motifs upstream of the CRISPR to fold 150 base-pairs of the genome for site-specific integration of foreign DNA.
Current Research Questions:
- How do other bacteria and archaea use DNA motifs and novel host factors to regulate CRISPR integration? How does this inform our general understanding of DNA mobilization (e.g., integration or transposition) in non-model organisms outside the phylum Pseudomonodota?
- The core CRISPR integrase Cas1-2 is commonly equipped with ancillary biochemical activities (e.g., nuclease, helicase, reverse transcriptase) throughout the phylogeny of CRISPR systems. How are these ancillary biochemical activities regulated, and what role do they play in the site-specific formation of DNA-based memories?
Mechanisms and applications of CRISPR-associated immune signaling pathways
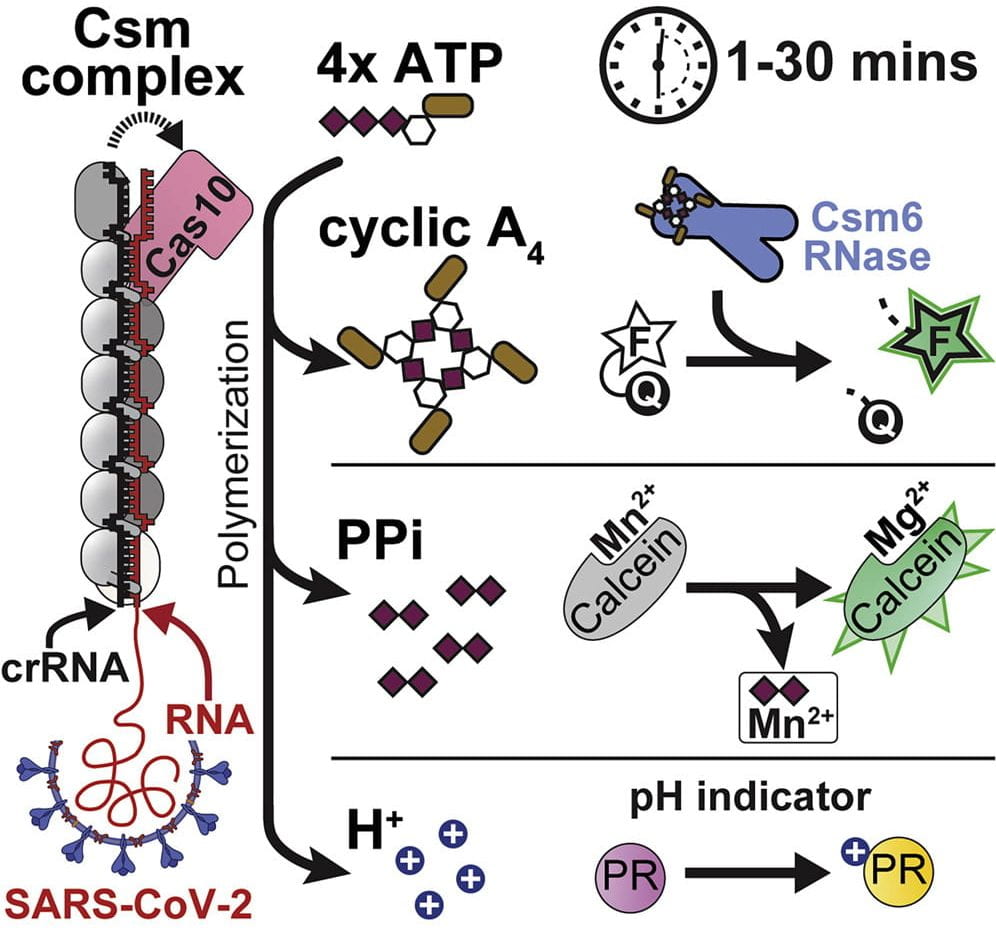
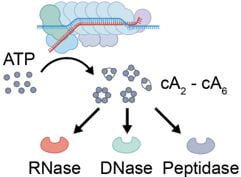
Nucleotides are the most primitive cellular messengers that regulate critical processes across the tree of life, including anti-viral immune responses, cell morphology, and motility. Recent studies show eukaryotic innate immune systems such as cGAS-STING share ancestry with bacterial immune systems. For example, viral RNA-bound Csm and Cmr complexes generate nucleotide messengers (e.g., cA3) that abort infection (e.g., RNA or DNA degradation) by activating immune effector proteins. To address a growing need for rapid and sensitive diagnostics, I co-invented an innovative RNA-guided Csm system for sensitive and sequence-specific detection of SARS-CoV-2 RNA, that repurposes a nuclease immune effector.
Computational “guilt-by-association” analyses suggest that CRISPR-generated immune signaling molecules activate a broad range of effectors, and kick-off diverse biochemical cascades – Some of these immune cascades bear a striking similarity to eukaryotic immune pathways.
Current Research Questions:
- How do diverse CRISPR-based signaling pathways trigger an immune response?
Funding:
- A.S-F. is an M. Jane Williams and Valerie Vargo Presidential Assistant Professor of Biology.
- Our research is currently supported by the NIH (R00GM147842*), and is Supported in part by the Institute for RNA Innovation of the Perelman School of Medicine at the University of Pennsylvania, and by the University Research Foundation at the University of Pennsylvania.
- A.S-F. has previously been supported by the NIH (K99GM147842), the Life Sciences Research Foundation, and the Simons Foundation, and Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund (G-1021106.01)
- S.A. is supported by the Frances Velay Fellowship Program, which is supported by funding from the Panaphil and Uphill Foundations.
- A.T. is supported by the Ruth Marcus Kanter College Alumni Society Undergraduate Research Grant.
- S.Y. is supported by Ernest M. Brown, Jr. College Alumni Society Undergraduate Research Grant.
- Undergraduate research is supported by Penn’s Center for Undergraduate Research & Fellowships, through the Grant for Faculty Mentoring Undergraduate Research award, and the Penn Undergraduate Research Mentoring program.
- Work in our lab also utilizes computational resources of the NIH STRIDES Initiative (https://cloud.nih.gov) through the Other Transaction agreement OT2OD027852.
*Note: Sadly, this funding was cancelled prematurely on April 2nd 2025. Reason: “due to changes in NIH/HHS priorities, the Maximizing Opportunity for Scientific and Academic Independent Careers (MOSAIC) program has been terminated.”